ChimeraX: Display AlphaFold predictions and error estimates using ChimeraX
Using ChimeraX, you can also visualize the predictions and PAE plots directly. This is done by ChimeraX by fetching data from the AlphaFoldDB (as described in this exercise), but you can also load in your own predictions.
PART 1: Fetch a predicted structure from AlphaFoldDB
A tutorial video for this exercise is available via: https://www.youtube.com/watch?v=oxblwn0_PMM.
- Open the AlphaFold window in the Chimera Gui via Tools > Structure Prediction > AlphaFold.
- Fetch an AlphaFold prediction from the Alphafold-EBI database by setting the Sequence input field to ‘UniProt identifier’ and provide the Uniprot ID of the protein of interest, e.g. INSR_HUMAN. The model is displayed and colored according to the pLDDT-score (confidence level).
- To show the associated PAE error plot click on Tools > Structure Prediction > AlphaFold Error Plot, enter the UniProt ID and click open. The color palette of the displayed PAE plot can be changed by clicking the right mouse button. E.g. ‘Color Plot green’ changes to the coloring scheme used on the Alphafold-EBI database.
- In the PAE plot, the distinct square-like regions along the diagonal with low PAE error values represent individual domains or continuous, rigid segments of the protein. Drawing a box in the PAE plot with the left mouse button will highlight the corresponding region in the atomic model.
- Off-diagonal regions with low PAE values indicate interacting regions or domains for which the relative position is accurately predicted. Drawing a box on these regions will highlight the two regions of interest in the atomic model.
- Clicking on ‘Color PAE domains’ segments the model in regions that are predicted to be rigid based on an analysis of the PAE plot (script written by Tristan Croll, https://github.com/tristanic/pae_to_domains). By inspecting the PAE plot you can assess if the segmentation makes sense.
(Optional) Steps via command-line:
AlphaFold prediction and errors can also be displayed using the ChimeraX command line.
- Fetch the prediction from the Alphafold-EBI database (
alphafold fetch p29474ORopen p29474 from alphafold) - Show the associated PAE error plot (
alphafold pae #1 uniprotId P29474 version 4) - Color the prediction according to the PAE domains (
alphafold pae #1 colorDomains true)
When working with Alphafold predictions that you generated yourself you can use the following command to color the model according to the pLDDT score:
color bfactor #1 palette alphafold
A detailed overview of the alphafold related commands in ChimeraX is available here: https://www.cgl.ucsf.edu/chimerax/docs/user/commands/alphafold.html
PART 2: Visualize your own prediction
To load in your own predictions into ChimeraX, you will need the predicted structure (.pdb file) and the PAE values, which are stored in the corresponding .pkl file.
- In ChimeraX, open the predicted structure. (for instance:
relaxed_model_2_multimer_v3_pred_0.pdb) - Go to Tools > Structure Prediction > AlphaFold Error Plot. ChimeraX will attempt to suggest the corresponding
.pklfile. In this case, the filenames are identical (except for the file extension). However, if you want to use the top ranked model, you should check theranking_debug.jsonfile to see which prediction corresponds to the top model!
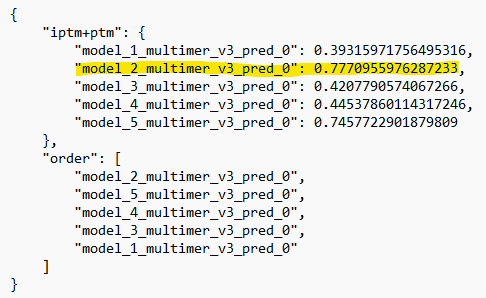
3. After confirming, the interactive PAE plot should be opened.